Publications (Google Scholar Profile)
Pre-prints
Peer-reviewed Publications
Characterization of the transcriptional divergence between the subspecies of cultivated rice (Oryza sativa) Campbell MT, Du Q, Liu K, Sharma S, Zhang C, Walia H. 2020. Characterization of the transcriptional divergence between the subspecies of cultivated rice (Oryza sativa). BMC Genomics. 21(1):1-6.
Leveraging genome-enabled growth models to study shoot growth responses to water deficit in rice Campbell M.T., Grondin A., Walia H., Morota G. (2020) Leveraging genome-enabled growth models to study shoot growth responses to water deficit in rice. J. Exp. Bot 71(18):5669–5679
Variance heterogeneity genome-wide mapping for cadmium in bread wheat reveals novel genomic loci and epistatic interactions Hussain W., Campbell M.T., Jarquin D., Walia H., Morota G. (2020) Variance heterogeneity genome-wide mapping for cadmium in bread wheat reveals novel genomic loci and epistatic interactions. The Plant Genome.
Utilizing trait networks and structural equation models as tools to interpret multi-trait genome-wide association studies. Momen, M., M.T. Campbell, Walia H, Morota G (2019). Plant Methods: 15(107).
In conventional multi-trait GWAS approaches the covariance between phenotypes are leveraged to improve the detection of genetic signal, however with these approaches we don’t know how the QTL affects the phenotype(s). Medhi used a Bayesian network approach to eludicate the relationship between biomass (shoot and root), water use, and water use efficiency. Structural equation modeling integrated with GWAS to decompose QTL effects and elucidate how a QTL acts on mutiple traits.
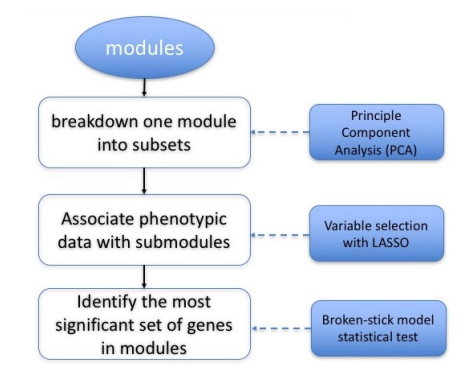
Using LASSO in gene co-expression network for genome-wide identification of gene interactions responding to salt stress in rice. Du, Q., M.T. Campbell, H. Yu, K. Liu, H. Walia, Q. Zhang, C. Zhang. (2019). Plant Direct (In Press).
Here, we used LASSO to select gene coexpression modules associated with salinity tolerance traits.
Predicting longitudinal traits derived from high-throughput phenomics in contrasting environments using genomic Legendre polynomials and B-splines. Momen, M., M.T. Campbell, Walia H, Morota G (2019). G3: Genes, Genomes, Genetics (In Press).
We sought to apply random regression models to forecast shoot growth trajectories using B-splines and Legendre polynomials in well-watered and water-limited conditions under various longitudinal cross-validation scenarios. We showed that the frequency of phenotypic evaluation can be reduced without impacting prediction accuracy.
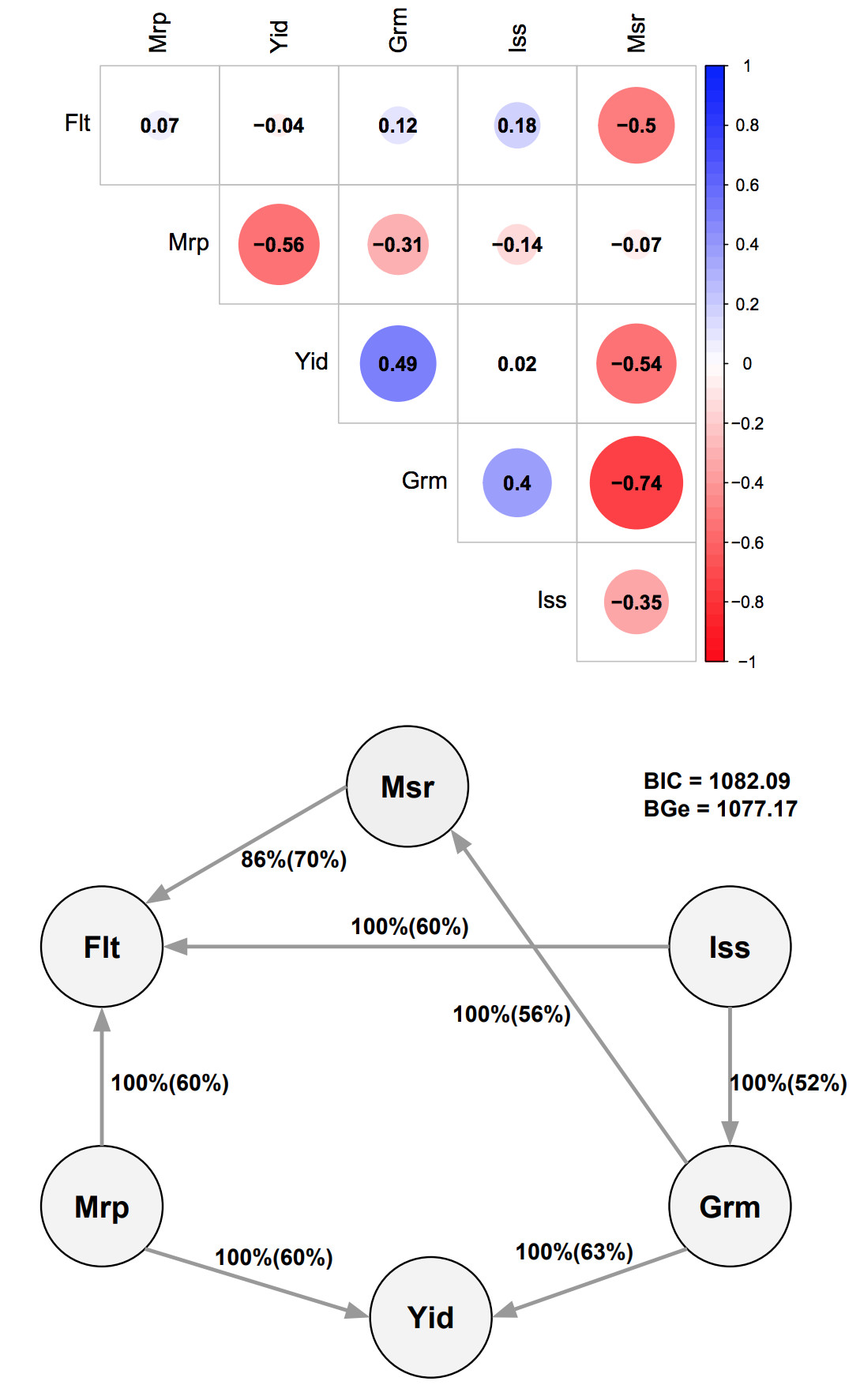
Genomic Bayesian confirmatory factor analysis and Bayesian network to characterize a wide spectrum of rice phenotypes. Yu, H., M.T. Campbell, Q. Zhang, H. Walia, G. Morota. (2019). G3: Genes, Genomes, Genetics.
This is a really nice approach to reduce the dimensionality of phenomics datasets and understand the genetic interrelationships between trait classes. Haipeng used confirmatory factor analysis to reduce 48 observed phenotypes into six latent variables, which essentailly respresent unobserved biological processes that contribute to the traits, and used Bayesian network to understand the interdependence among latent variables.
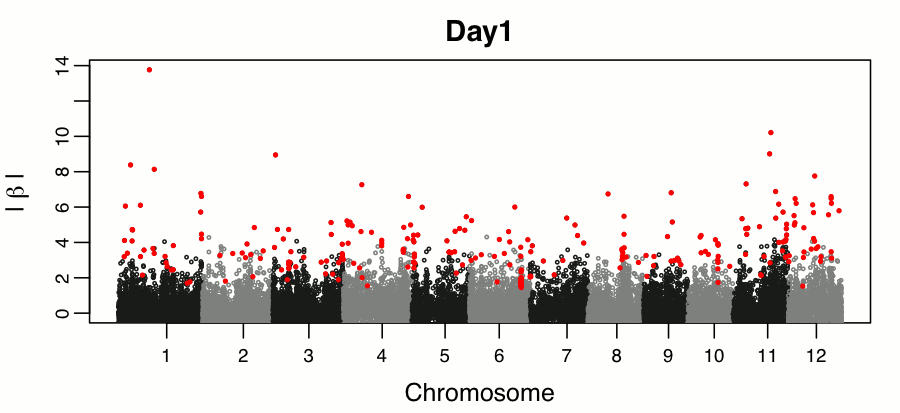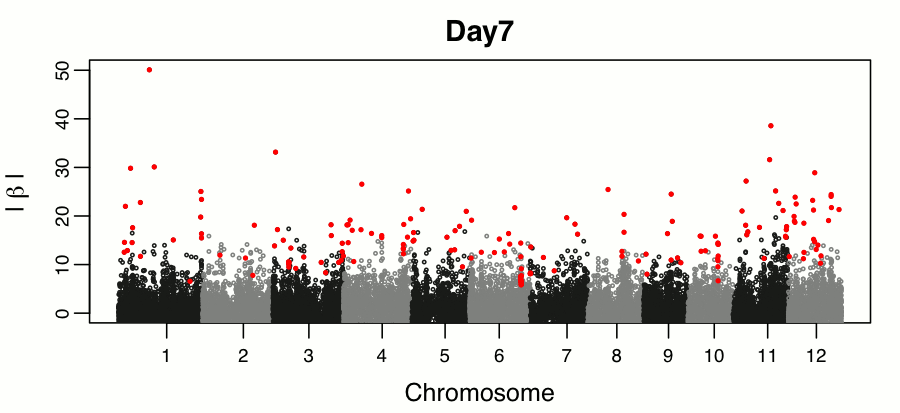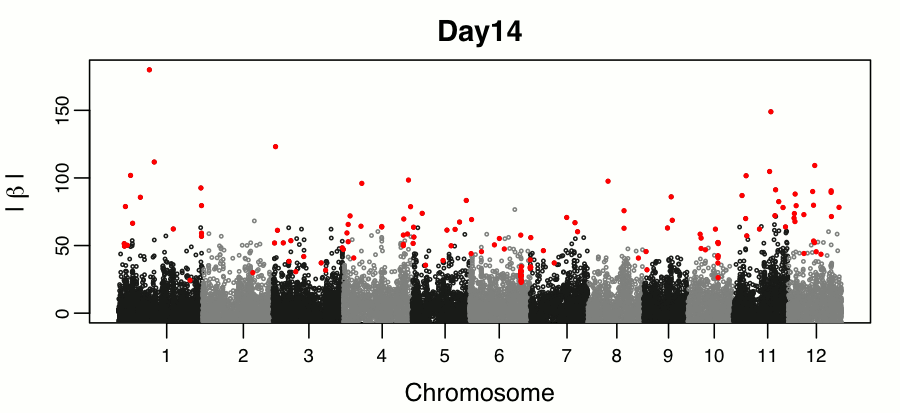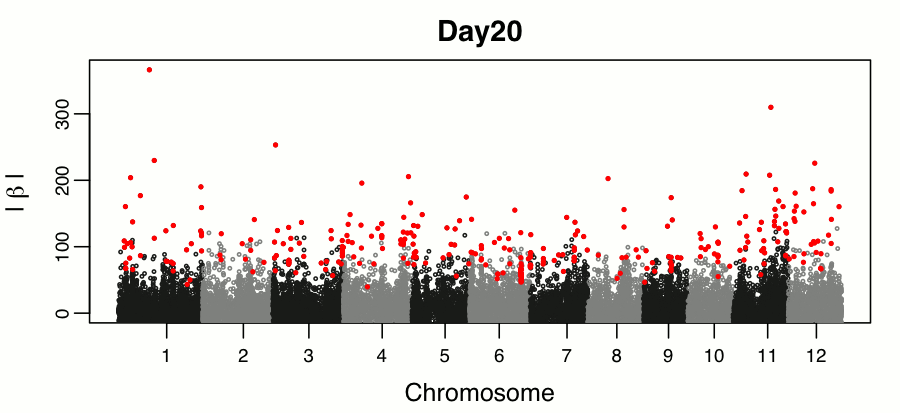
Leveraging breeding values obtained from random regression models for genetic inference of longitudinal traits. M.T. Campbell, M. Momen, H. Walia, G. Morota. (2019). The Plant Genome..
This study builds on the random regression genomic prediction approach described in Campbell et al 2018, and used the derived breeding values for genomic inferenece across time points.
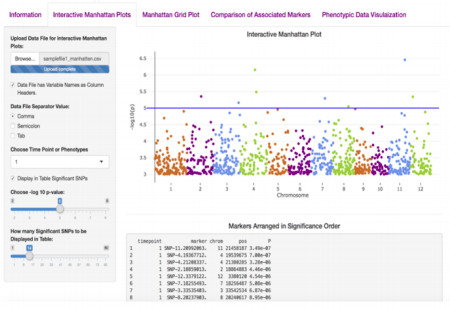
ShinyAIM: Shiny-based Application of Interactive Manhattan Plots for Longitudinal Genome-Wide Association Studies. Hussain, W., M.T. Campbell, H. Walia, G. Morota. (2018). Plant Direct.
This is a nice tool to easily visualize multiple manhattan plots. Wasseem packaged a bunch of useful methods to explore the phenotypic data and GWAS results into this app. This is particularly useful for phenomics datasets or longitudinal studies.
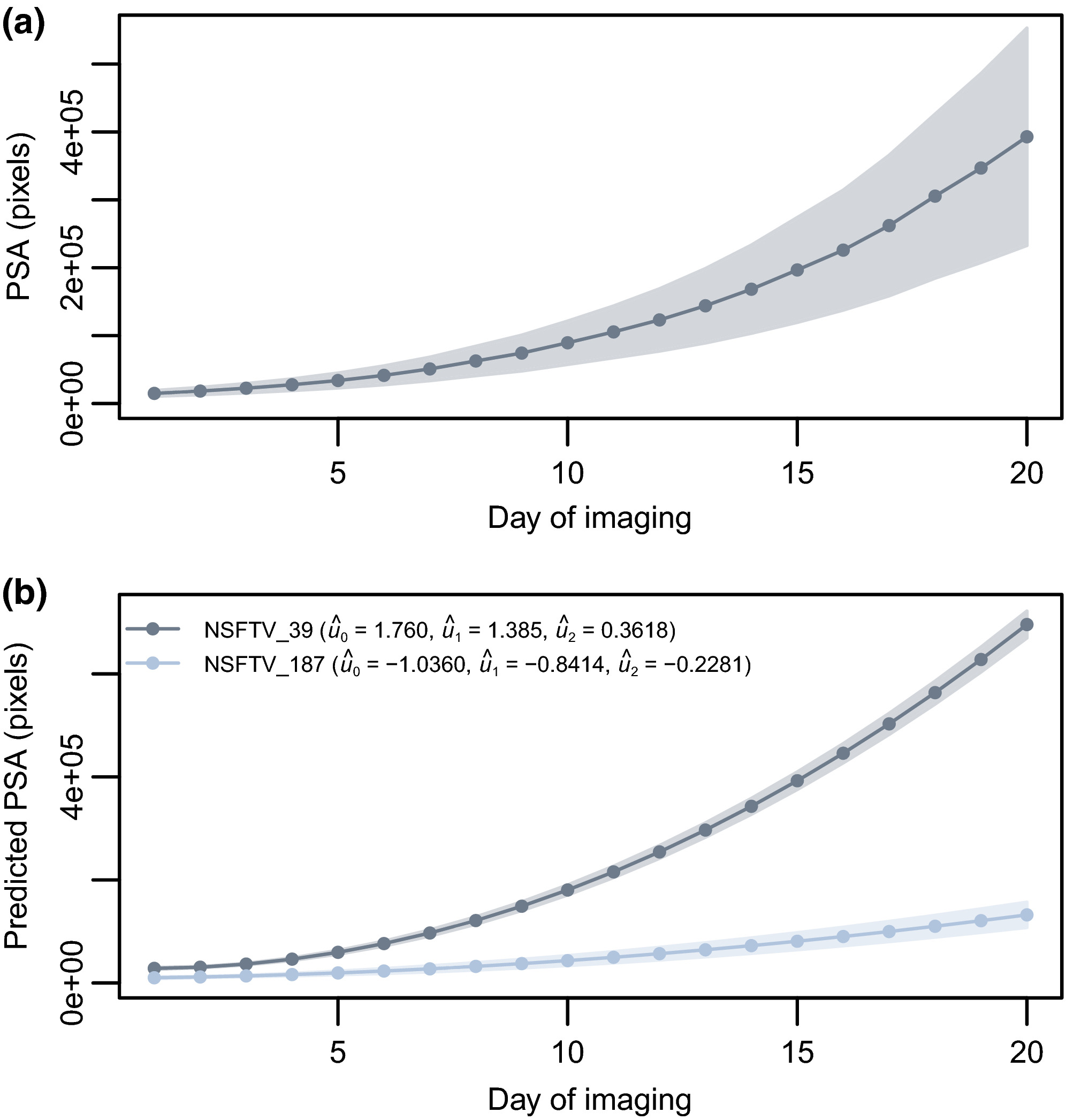
Utilizing random regression models for genomic prediction of a longitudinal trait derived from high-throughput phenotyping. Campbell, M.T., H. Walia, and G. Morota. (2018). Plant Direct.
This paper utilized a random regression approach for genomic prediction of longitudinal shoot growth trajectories in rice. The RR approach sigificant imporved prediction accuracies compare to a single time point approach. To our knowledge, this is the first study to implement this approach in a crop species.
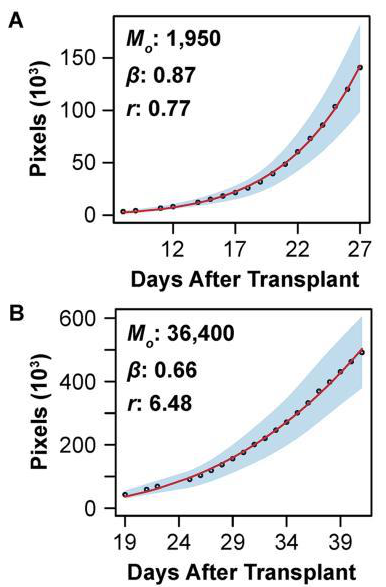
A Comprehensive Image based Phenomic Analysis Reveals the Complex Genetic Architecture of Shoot Growth Dynamics in Rice (Oryza sativa). Campbell, M.T. , Q. Du, K. Liu, C.J. Brien, B. Berger, C. Zhang, and H. Walia. (2017). Plant Genome.
We used a “two-step” functional GWAS and genomic prediction approach to study shoot growth dyanamics in rice. First, we modeled shoot biomass over 20 days using a power law function and used model parameters as derived traits for association mapping and genomic prediction.
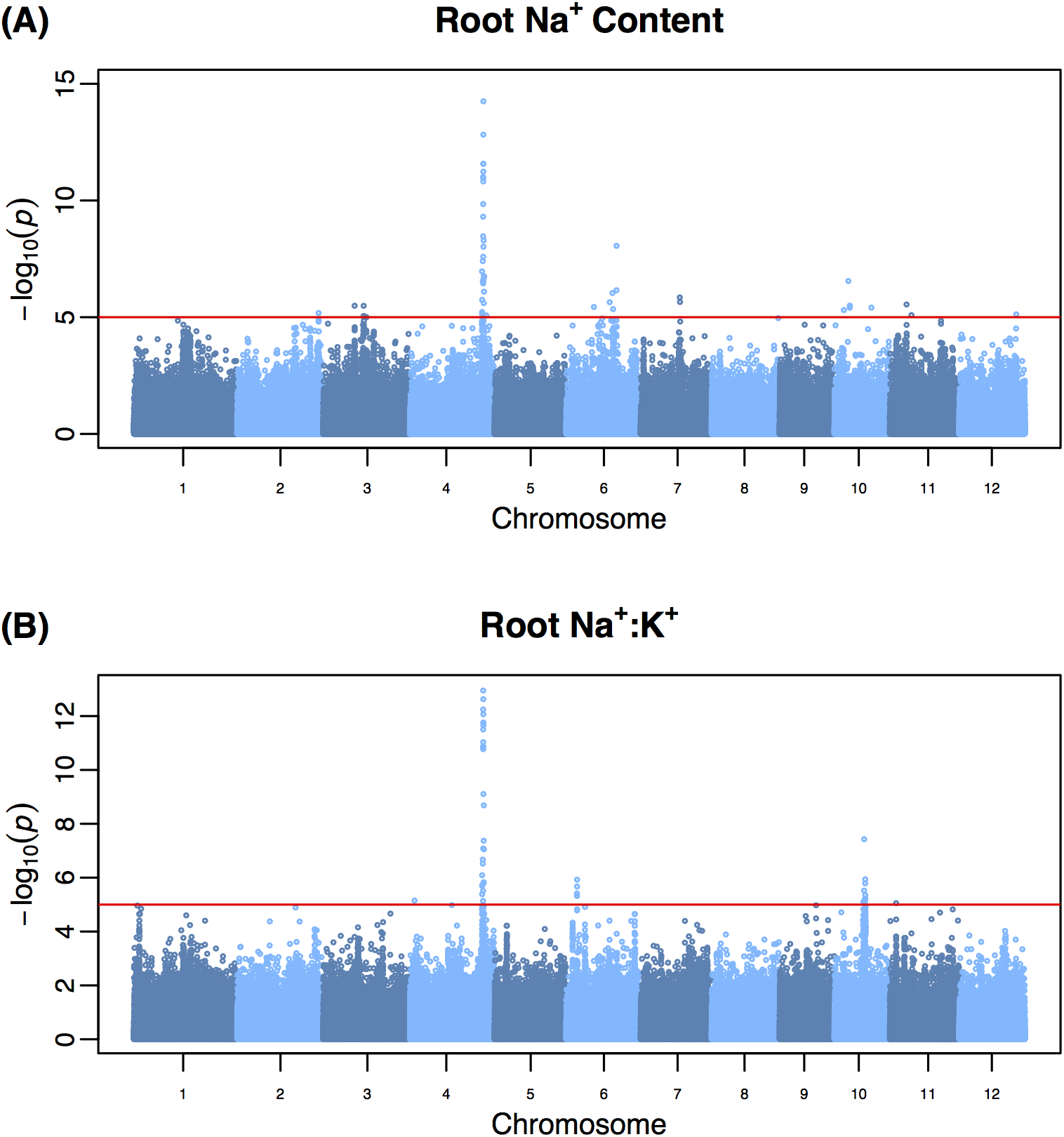
Allelic variants of OsHKT1;1 underlie the divergence between indica and japonica subspecies of rice (Oryza sativa) for root sodium content. Campbell, M.T. , N. Bandillo, F.R.A. Al Shiblawi, S. Sharma, K. Liu, Q. Du, A.J. Schmitz, C. Zhang, A.A. Very, A.J. Lorenz, and H. Walia. (2017). PLoS Genetics.
HKT1;1 was identified as a major regulator of root sodium content in rice. Variants that influence the transport of sodium were found to be a component of the divergence between to two subspecies of rice for root sodium content.
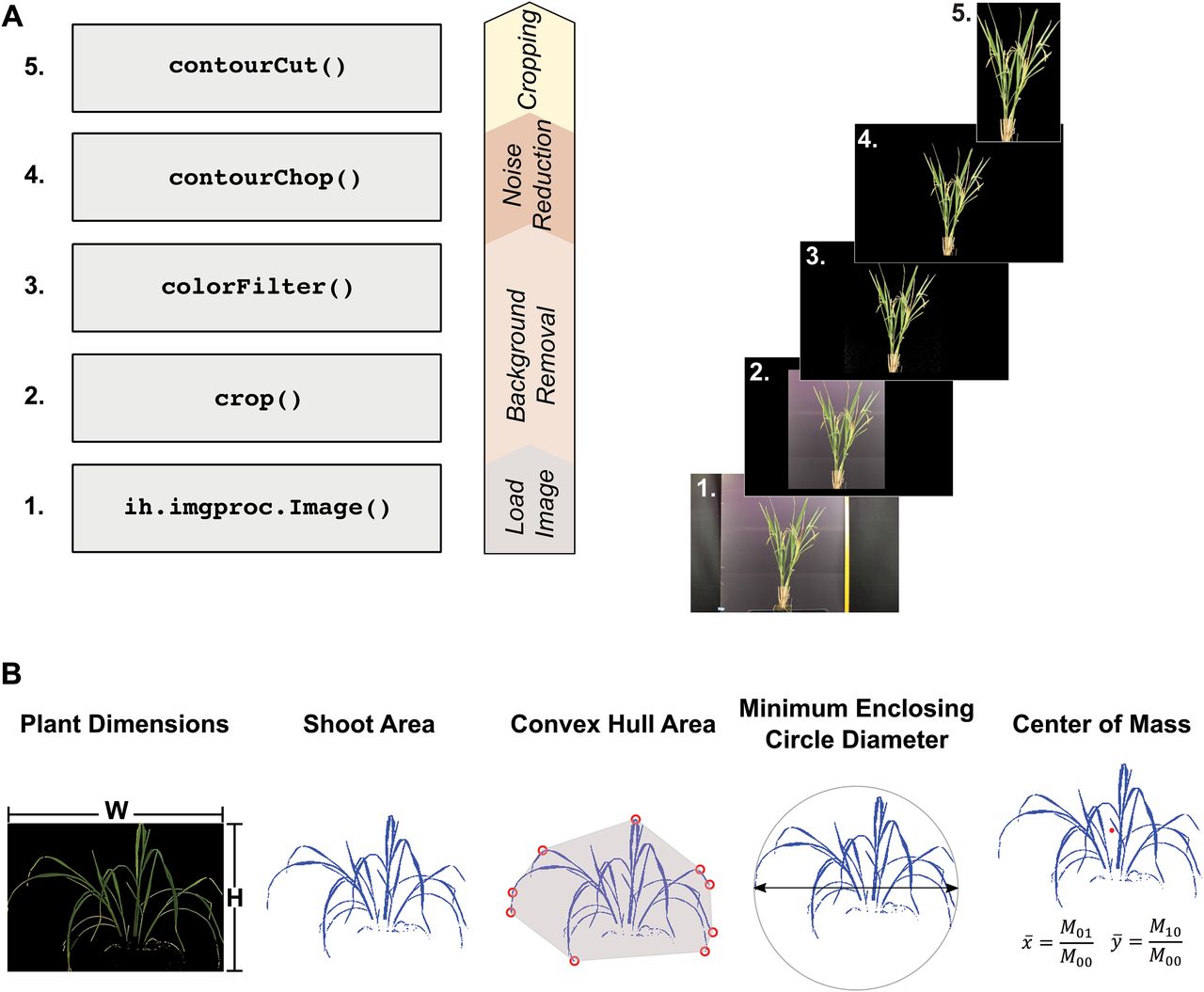
Image Harvest: an open source platform for high throughput plant image processing and analysis. Knecht, A.C.†‡, M.T. Campbell† , A. Caprez, D.R. Swanson, and H. Walia. (2016). Journal of Experimental Botany.
We developed a software to process and anayze images derived from high throughput phenotyping platforms.
† Equal contribution
‡ Undergraduate coauthor
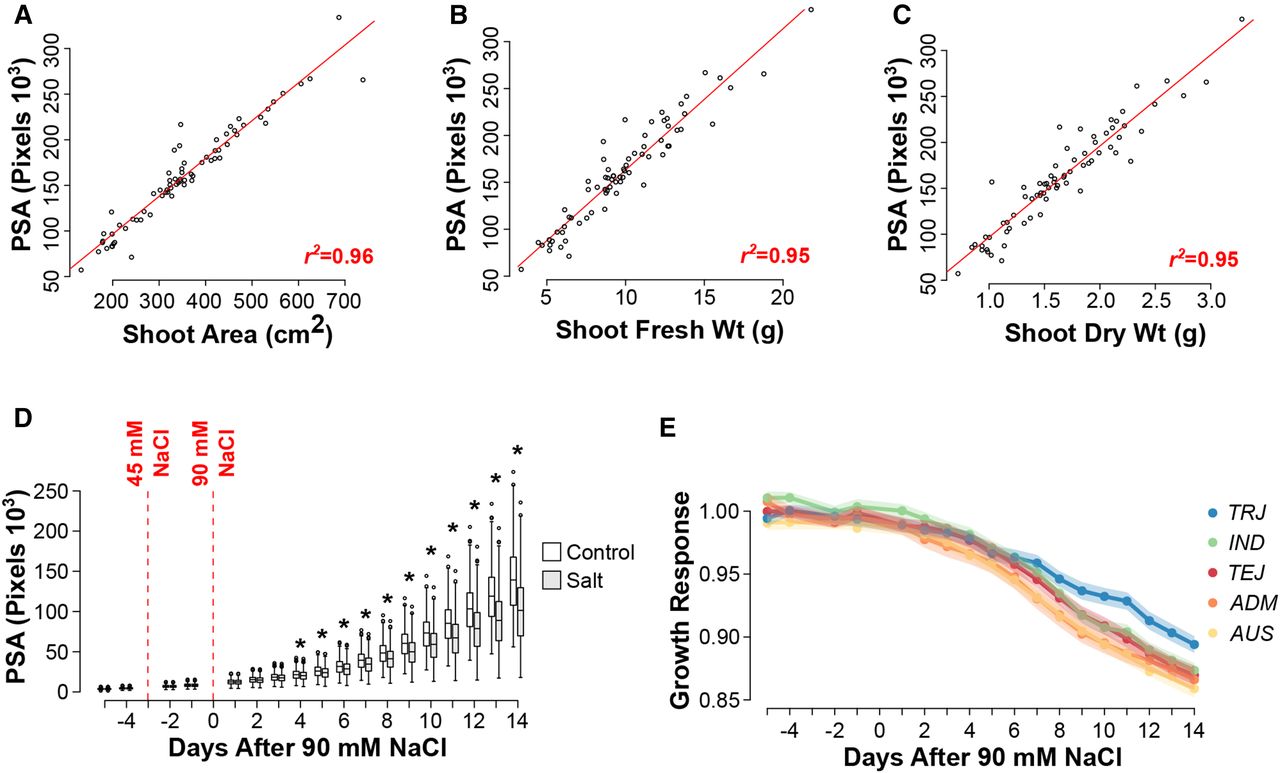
Integrating image based phenomics and association analysis to dissect the genetic architecture of temporal salinity responses in rice. Campbell, M.T., A.C. Knecht, B. Berger, C.J. Brien, D. Wang, and H. Walia. (2015). Plant Physiology.
Here, we used image based high thoughput phenotyping and association mapping to examine the genetic basis of temporal salinity responses.
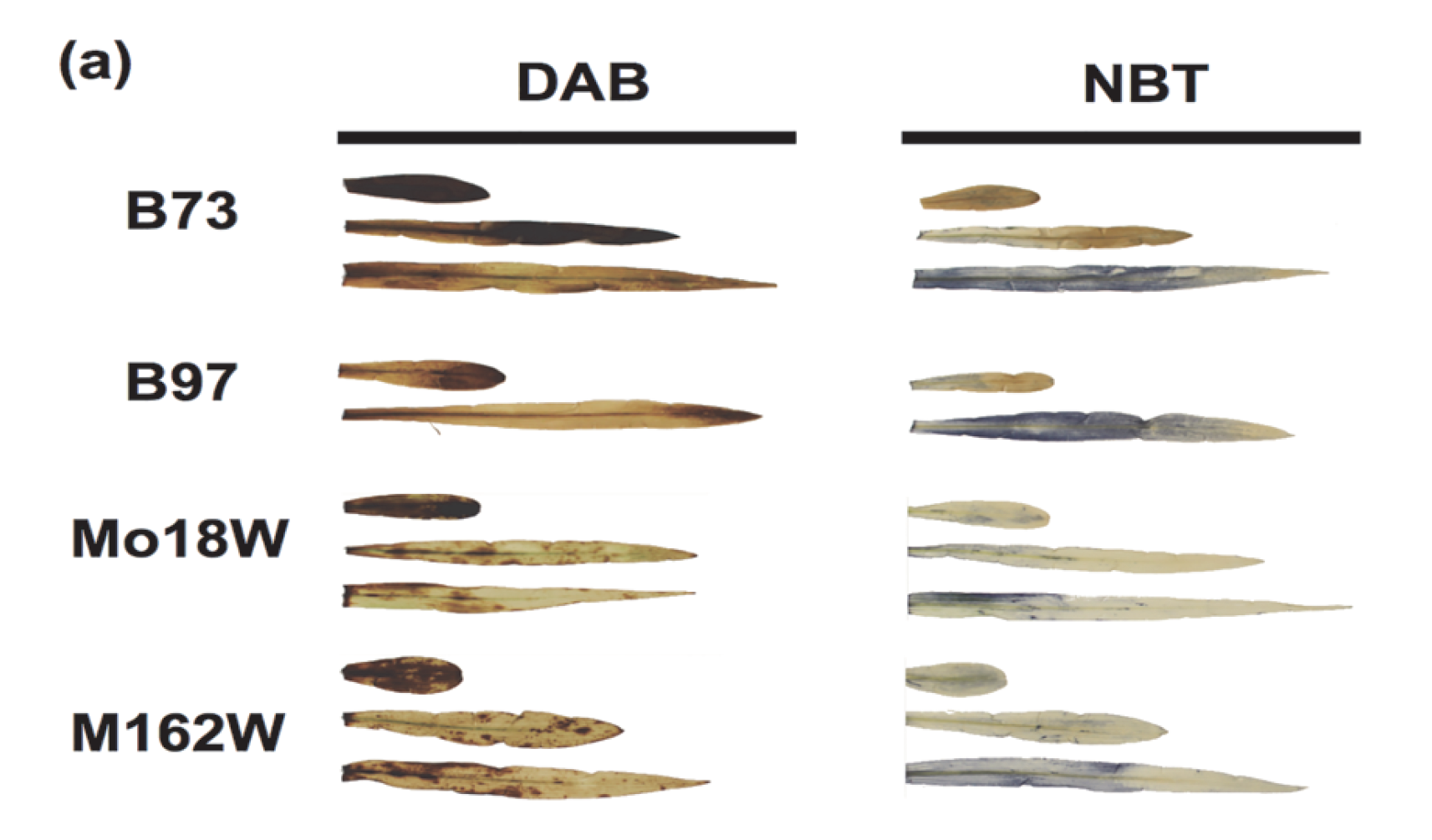
Genetic and Molecular Characterization of Submergence Response Identifies Subtol6 as a Major Submergence Tolerance Locus in Maize. Campbell, M.T., Campbell, M.T., C.A. Proctor, Y. Dou, A.J. Schmitz, P. Phansak, G.R. Kruger, C. Zhang, and H. Walia. (2015). PLoS One.
This paper assessed natural variation for submegence tolerance in maize and identifies a QTL that influences submegence tolerance.
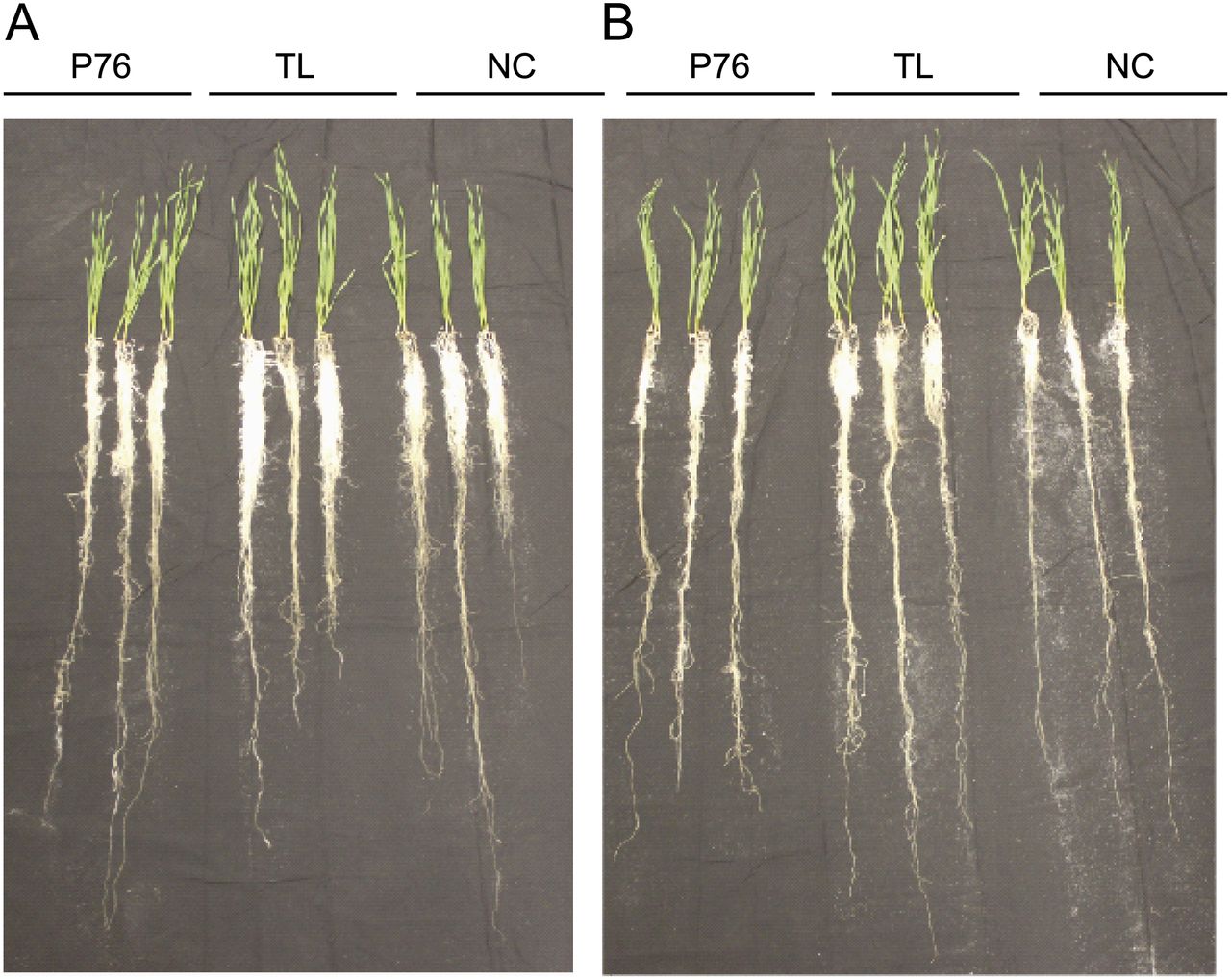
Introgression of Novel Traits from a Wild Wheat Relative Improves Drought Adaptation in Wheat. Placido, D.F., M.T. Campbell, J.J. Folsom, X. Cui, G.R. Kruger, P.S. Baenziger, and H. Walia. (2013). Plant Physiology.
This paper looks at the effects of an introgression from Agropyron elongatum into Triticum aestivum on drought tolerance.
